4th European Meeting on Planarian Biology

SEMINAR: Peter Dearden (Univ Otago, NZ)
SEMINAR
Wednesday, 20 July, 11:00
Neil Chalmers Theatre, DC2
Peter Dearden
Director of Genetics Otago & Associate Dean of Research for Health Sciences
University of Otago, New Zealand

Notch signalling has been co-opted by queen honeybees to repress worker reproduction
Eusociality depends on reproductive division of labour. This implies that one individual, the queen in honeybees, can repress reproduction in other females. In honeybees this is achieved through queen bees producing a pheromone, queen mandibular pheromone, which causes worker bees to repress the activity of their ovaries, repressing worker reproduction. Removal of the queen, and thus queen mandibular pheromone, allows the workers to activate their ovaries and begin to lay eggs. We have examined changes in gene expression, using high-throughput sequencing, in worker ovaries as they respond to the absence of queen mandibular pheromone and become activated. Using this dataset we have investigated the mechanisms by which queen mandibular pheromone has its action. While it is not clear how queen mandibular pheromone is detected, or how its activity is transduced, we have identified a cell-signalling pathway that responds to the absence of queen pheromone, and, when blocked, enhances the activation of the ovaries. Comparison of the action of this pathway in the presence or absence of queen pheromone implies that this is the main pathway by which the activity of worker ovaries is constrained in the honeybee, giving us insights into both the mechanism and evolution of reproductive constraint.
Who's going to save the tapeworms?

Read Jason Bittel's expose on the loss of the unique New Zealand kakapo, and with it, it's unique parasite fauna, including the tapeworm Stringopotaenia psittacea
ISOMORPHOLOGY: An introduction to the art and science of Gemma Anderson
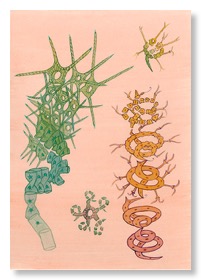
Gemma Anderson is unusually gifted artist who works at the interface of art, science and philosophy. In a recent blog post of the Journal of Zoology Gemma introduces her concept of 'isomorphology': an approach to discovering and understanding patterns and processes in the natural world through the process of drawing. Gemma has worked in association with a broad range of scientists, scientific fields, and scientific institutions and we have been privileged to host her in our lab where she has applied her unique skills to help us understand the complex ontogeny of tapeworms. Gemma is currently working on a funding application that will allow her to collaborate with a range of labs interested in developmental processes from the cell to the whole organism, and we look forward to continuing her association with our lab.
For more information please visit Gemma's WEBSITE
British Society of Parasitology, 11-13 April 2016
 Join us at Imperial College, London, for the British Society of Parasitology where Pete and Francesca will make the following presentations:
Join us at Imperial College, London, for the British Society of Parasitology where Pete and Francesca will make the following presentations:Thursday, April 13 - 2:45-3:00
LT 340 - Huxley Building
Session: Helminth Signalling and Developmental Parasitology I
From planarians to parasitism: Wnt/Hedgehog signalling controls AP patterning during larval and strobilar development in tapeworms
Francesca Jarero*, Uriel Koziol & Peter D. Olson
Wnt/Hedgehog signalling in free-living planarians is responsible for mediating head/tail decision making during early development and in regeneration. More broadly, canonical Wnt signalling has been found to underlie AP axis formation in animals generally. We show that AP specification during larval metamorphosis in tapeworms also involves canonical Wnt signalling, with scolex formation taking place at the site(s) of Wnt repression. Moreover, we show that the same system has been co-opted during strobilar development, with segmental boundaries expressing opposing ‘stripes’ of anteriorizing (SFRP) and posteriorizing (Wnt1) signals—tapeworm strobilation therefore being a form of paratomy. Expression of Hedgehog and Hox also appear linked to the system, upstream and downstream of Wnt signaling, mirroring the model of AP signalling in planarians as presently understood. Taken together, we show that parasitic and free-living flatworms share the same underlying AP patterning system despite their highly disparate body plans.
Thursday, April 13 - 3:45-4:15
LT 340 - Huxley Building
Session: Helminth Signalling and Developmental Parasitology II
Tapeworm tumours: how not to make a tapeworm
Peter D. Olson* & Atis Muehlenbachs
In a recently reported case of dwarf tapeworm (Hymenolepis nana) infection in an HIV-positive Columbian man (NEJM 373:1845-52), tumours were found in the lymph nodes, lungs and elsewhere that appeared to comprise non-human cells. Genetic analyses confirmed these to be H. nana cells, and their small size (< 10 um), spherical shape and low cytoplasm/nuclear ratio were all consistent with tapeworm germinative cells or ‘neoblasts' (i.e. flatworm stem cells). With the exception of a degree of syncitia formation (characteristic of tapeworm tissue architecture generally) no cell differentiation was seen. However clonal proliferation and evasive dissemination was evidenced by genetic analysis and the tumours proved fatal. Preliminary genomic analysis comparing the case study with a reference laboratory isolate revealed breaks in genes previously implicated in human cancers (e.g. LAMP2). I describe this case together with previous cases of tapeworm infection in immunocompromised patients in order to illustrate the most likely route of infection and demonstrate the necessity for cross-talk with the host’s immune system for normal development of the parasite. Lastly, I will briefly outline why studying parasites as animals rather than as some unique category of being is essential for understanding their biology and informs applied areas of NTD research.
Next Generation Systematics

We live in an age of ubiquitous genomics. Next generation sequencing (NGS) technology, both widely adopted today and advancing at pace, has transformed the data landscape, opening up an enormous source of heritable characters to the comparative biologist. Its impact on systematics, like many other fields of biology, has been felt throughout its breadth: from defining species boundaries to estimating their evolutionary histories.
This volume examines the broad range of ways in which NGS data are being used in systematics and in the fields that it underpins, from biodiversity prospecting to evo-devo. The authors draw on contemporary case studies to demonstrate state-of-the-art applications of NGS data. These, along with novel analyses, comprehensive reviews and lively perspectives are combined to produce an authoritative account of contemporary issues in systematics that have been advanced and impacted by the recent adoption of NGS.
Peter D. Olson is a Researcher in the Department of Life Sciences at the Natural History Museum, London. His research integrates comparative, developmental and genomic studies of parasitic flatworms aimed at unravelling the genetic basis of their evolution. He became involved in the field of molecular phylogenetics when PCR and manual sequencing were being widely used to generate the first ribosomal-based estimates of animal phylogeny.
Joseph Hughes is an evolutionary biologist working at the Medical Research Council – University of Glasgow Centre for Virus Research. He is particularly interested in understanding the evolutionary forces that have shaped the solutions that different species have found to adapt to their environment. He has embraced the deluge of data from high throughput sequencing platforms and currently works on non-living particles known as viruses.
James A. Cotton is a Senior Staff Scientist in the Parasite Genomics group at the Wellcome Trust Sanger Institute. His background is in phylogenetic theory and methods, and he now leads projects on comparative, evolutionary and population genomics of parasites of medical and veterinary importance.
The Systematics Association Special
Volume 85
Next Generation Systematics
Edited by
Peter D. Olson
Natural History Museum, London
Joseph Hughes
University of Glasgow
James A. Cotton
Wellcome Trust Sanger Institute, Cambridge
Contents
List of contributors
Introduction: studying diversity in an age of ubiquitous genomics
James A. Cotton and Peter D. Olson
Part I Next generation phylogenetics
1 Perspective: Challenges in assembling the ‘next generation’ tree of life
Michael J. Sanderson
2 The role of next-generation sequencing technologies in shaping the future of insect molecular systematics
Joseph Hughes and Stuart Longhorn
3 Phylogenomics of Nematoda
Mark Blaxter, Georgios Koutsovoulos, Martin Jones, Sujai Kumar and Ben Elsworth
4 High throughput multiplexed mitogenomics for metazoans—prospects and limitations
Peter G. Foster, Maria A. Stalteri, Andrea Waeschenbach and D. Timothy J. Littlewood
5 Investigating bacterial microevolution through next-generation sequencing
Josephine M. Bryant and Simon R. Harris
Part II Next generation biodiversity science
6 Perspective: Après le déluge: ubiquitous field barcoding should drive 21st century taxonomy
Richard M. Bateman
7 Perspective: Biodiversity and the (data) beast
Holly M. Bik and W. Kelley Thomas
8 Next generation biodiversity analysis
Mehrdad Hajibabaei and Ian King
9 Protist systematics, ecology and next generation sequencing
David Bass and Thomas Bell
Part III Next generation challenges and questions
10 Perspective: Systematics in the age of genomics
Antonis Rokas
11 Perspective: The role of next generation sequencing for integrative approaches to evolutionary biology
Ralf J. Sommer
12 Next-generation apomorphy: the ubiquity of taxonomically restricted genes
Paul A. Nelson and Richard J. A. Buggs
13 Utilizing next generation sequencing for evo-devo study of plant traits
Rachel H. Walker, Paula J. Rudall and Beverley J. Glover
14 An NGS approach to archaeobotanical museum specimens as genetic resources in systematics research
Oliver Smith, Sarah A. Palmer, Rafal Gutaker and Robin G. Allaby
15 From sequence reads to evolutionary inferences
James A. Cotton
Index